- Definition: Natural Language Processing (NLP) is the study of the computational treatment of natural (human) language, i.e., teaching computer how to understand and generate human language.
- Research Resource: NLP draws on research in
- Linguistics
- Theoretical Computer Science
- Mathematics
- Statistics
- Artificial Intelligence
- Psychology
- Database
- Language and Communication (Speaker):
- Intention- goals, shared knowledge and beliefs
- Generation- tactical
- Synthesis- text or speech
- Language and Communication (Listener):
- Perception
- Interpretation- syntactic, semantic, pragmatic
- Incorporation- internalization. understanding
- Basic NLP Pipeline: Language --(Understanding)--> Computer --(Generation)--> Language
- Challenges: coming from the text
- current events, background events, speculation, property, reference to previous sentence
- genres of text: blogs, emails, press releases, chats, debates, etc.
- special types of text and terminologies
- incomplete sentences
- fiction text, rare words,
- extremely long and complex sentences
- multiple possible interpretations, ambiguous sentences
- lexical, structural, scope ambiguity
- Textbook
- Speech and Language Processing (3rd edition) (Dan Jurafsky and James H. Martin) or 2nd edition
- Foundations of Statistical Natural Language Processing (Chris Manning and Hinrich Schütze)
- Natural Language Understanding (James Allen)
- Courses
- JHU (Jason Eisner)
- Cornell (Lillian Lee)
- Stanford (Chris Manning)
- U. Maryland (Hal Daume)
- Berkeley (Dan Klein)
- U. Texas (Ray Mooney)
- Coursera (Manning/Jurafsky, survey)
- Coursera (Michael Collins, advanced, 2013)
- Related Fields
- CL (Computational Linguistics)- more mathematical and formal treatment of linguistics, less confusing on applications
- IR (Information Retrieval)- study of finding information and documents (text, speech or video).
- SP (Speech Processing)- deal with understanding and generation of spoken signals.
- HLT ( Human Language Technology)- uses of the applied component of the NLP.
- NLE (Natural Language Engineering)- synonym of HLT.
- ML (Machine Learning)- computational, statistical study of learning.
- Research in NLP
- Conference: ACL/NAACL, EMNLP, SIGIR, AAAI/IJCAI, Coling, HLT, EACL/NAACL, AMTA/MT Summit, ICSLP/Eurospeech
- Journals: Computational Linguistics, TACL, Natural Language Engineering, Information Retrieval, Information Processing and Management, ACM Trans. on Information Systems, ACM TALIP, ACM TSLP
- ACL Anthology
- ACL Anthology Network (AAN)
- Difficulties of NLP
- Similar forms does not imply similar meanings (e.g., "Beverly Hills/Sills")
- Computer is not good at metaphors (e.g., "Time flies like an arrow.")
- Word order is important (e.g., "The box/pen is in the pen/box.")
- Not the same semantic relationship (e.g., Mary and Sue are mothers/sisters.")
- Not the same order of magnitude due to semantic change (e.g., "Every American has a mother/president.")
- Draw on common sense (e.g., "We gave monkeys bananas because they were hungry/over-ripe.")
- Syntactic correctness does not guarantee semantic correctness (e.g., "Colorless green ideas sleep furiously.")
- Ambiguous words due to multiple meanings (e.g., "ball"), part of speech (e.g., "fly"), different pronunciations (e.g., "address"), noun-noun phrases (e.g., "Science fiction writer"), etc.
- Types of Ambiguity
- Morphological- "Joe is quite impossible/important."
- Phonetic- "Joe's finger got number."
- Part of Speech- "Joe won the first round."
- Syntactic- "Call Joe a taxi."
- Pp Attachment- "Joe ate pizza with a for/with meatballs/with Sam/with pleasure."
- Sense- "Joe took the bar exam."
- Modality- "Joe may win the lottery."
- Subjectivity- "Joe believes the stocks will rise."
- Cc Attachment- "Joe likes ripe apples and pears."
- Negation- "Joe likes his pizza with no cheese and tomatoes."
- Referential- "Joe yelled at Mike. He had broken the bike." "Joe yelled at Mike. He was angry at him."
- Reflexive- "John brought him/himself a present."
- Ellipsis and Parallelism- "Joe gave Mike a beer and James a glass of wine."
- Metonymy- "Boston called and left a message for Joe."
- Other Difficulties
- Non-standard- "+1-212-772-1220", "A360"
- Slang- "friend (verb)", "spam"
- Novel words and usages- "yolo", "selfie"
- Inconsistencies- "Junior college" vs. "college junior"
- Typos and grammatical errors
- Parsing problems- "cup holder", "Federal Reserve Board Chairman"
- Complex sentences
- Counterfactual sentences- " if you were to do..."
- Humor and sarcasm
- Implicature/inference/world knowledge- "I was late because my car broken down." implies I have a car, I used the car to get to places, etc.
- Semantics vs. pragmatics- "Do you know the time?"
- Language is even hard for human (both L1 and L2).
- Synonyms and Paraphrases
- Synonyms- "climbed", "gained" and "rose"
- Paraphrases- "its best close", "for its best showing" and "its highest level"
- Linguistics Knowledge
- Constituents- "My cousin's neighbor's children eat pizza." and "Eat pizza"
- Colocations- "strong beer" but *"powerful beer"
- Get Linguistics Knowledge into the System
- Manual rules
- Automatically acquired from large text collections (corpora)
Bioinformatics
A multi-dimensional space between biology and computer science......
Friday, July 8, 2016
Natural Language Processing (NLP) Notes
This is the working notes for the Natural Language Processing (NLP) that is part of the bioinformatics project.
Monday, April 25, 2016
Today: National DNA Day!
Today, April 25, is the National DNA Day! The day commemorates the completion of the Human Genome Project (see PBS Nova video: Cracking the Code Of Life) in April 2003, and the discovery of DNA's double helix molecular structure in 1953 by James Watson and Francis
Crick.
63 years later, DNA analysis
and manipulation play major roles in nearly every aspect of our lives. It opens the door for new possibilities in medical field such as preventive medicine,  pharmacogenomics, and gene therapy. With the help of computer science and technology (Bioinformatics), gene sequencing has accelerated its speed exponentially, and it's cost has also drop exponentially
(even faster than the Moore's Law!). The result is that the available
genomics data has grown at the rate faster than any research can handle.
pharmacogenomics, and gene therapy. With the help of computer science and technology (Bioinformatics), gene sequencing has accelerated its speed exponentially, and it's cost has also drop exponentially
(even faster than the Moore's Law!). The result is that the available
genomics data has grown at the rate faster than any research can handle.
 pharmacogenomics, and gene therapy. With the help of computer science and technology (Bioinformatics), gene sequencing has accelerated its speed exponentially, and it's cost has also drop exponentially
(even faster than the Moore's Law!). The result is that the available
genomics data has grown at the rate faster than any research can handle.
pharmacogenomics, and gene therapy. With the help of computer science and technology (Bioinformatics), gene sequencing has accelerated its speed exponentially, and it's cost has also drop exponentially
(even faster than the Moore's Law!). The result is that the available
genomics data has grown at the rate faster than any research can handle.
With this big amount of data, machine learning will be the best candidate technology to tackle this challenge. Machine learning has brought breakthroughs in computer vision, natural language procession, data mining in the past few years. Now, it will become a powerful tool in bioinformatics to revolutionize the field of biology and medicine. While celebrating the National DNA Day, let's start our adventure!
Thursday, April 14, 2016
Machine Learning Applications in Biology
I visited Professor Weigang Qiu at Hunter College last Wednesday. He is leading the Evolutionary Bioinformatics Lab and his research is focusing on comparative analysis of multiple genomes of the Lyme disease pathogen. We discussed the possibility to run a research project to support high-school students conducting advanced research in bioinformatics field. I also explained to him our machine learning-related
activities, and our intention to expand our research activities into
the biological science. He believed that we can run projects focusing on
"machine-learning applications in biology". The possible applications may include gene prediction, cancer type classification based on gene expression, and gene function prediction (e.g., Borrelia, Lyme pathogen lipoproteins). Professor Qiu will explore more in this area and come up more detailed project description for us.
In the mean time, our focus will be in three aspects:
- Biology: Please review/preview chapters in your Biology/AP Biology textbook related to molecular genetics, gene expression, and genomes. If you need an AP Biology textbook, I can get one for you.
- Machine Learning: If you have not trained to understand the perceptron at code level, please arrange time to do it.
- Bioinformatics: You also encouraged to take any MOOC bioinformatics course in the last post to build up the basic programming skill and knowledge of bioinformatics.
Wednesday, March 30, 2016
Bioinformatics MOOC Courses
There are a few free bioinformatics MOOC courses:
Coursera:

EdX:
Coursera:
- Biology Meets Programming: Bioinformatics for Beginners,
- Bioinformatics: Introduction and Methods,
- Genomic Data Science 1: Introduction to Genomic Technologies,
- Genomic Data Science 2: Genomic Data Science with Galaxy,
- Genomic Data Science 3: Python for Genomic Data Science,
- Genomic Data Science 4: Algorithms for DNA Sequencing,
- Genomic Data Science 5: Command Line Tools for Genomic Data Science,
- Genomic Data Science 6: Bioconductor for Genomic Data Science,
- Genomic Data Science 7: Statistics for Genomic Data Science, and
- Genomic Data Science 8: Genomic Data Science Capstone.

EdX:
- Data Analysis for Life Sciences 1: Statistics and R,
- Data Analysis for Life Sciences 2: Introduction to Linear Models and Matrix Algebra,
- Data Analysis for Life Sciences 3: Statistical Inference and Modeling for High-throughput Experiments,
- Data Analysis for Life Sciences 4: High-Dimensional Data Analysis,
- Data Analysis for Life Sciences 5: Introduction to Bioconductor: Annotation and Analysis of Genomes and Genomic Assays,
- Data Analysis for Life Sciences 6: High-performance Computing for Reproducible Genomics, and
- Data Analysis for Life Sciences 7: Case Studies in Functional Genomics.
When Biology Meets Computer Science
Here is my
on-going study notes at the entry point of this fascinating multi-dimensional
space - Bioinformatics.

- Genome: a full set of chromosomes; all the inheritable traits of an organism.
- DNA: Deoxyribonucleic acid (DNA) is a molecule that carries most of the genetic instructions used in the development, functioning and reproduction of all known living organisms and many viruses. Most DNA molecules consist of two biopolymer strands coiled around each other to form a double helix.
- Nucleic Acid: DNA (along with RNA) is a nucleic acid; alongside carbohydrates, lipids, proteins, and nucleic acids compose the four major macromolecules essential for all known forms of life.
- Nucleotide: DNA is a long polymer made from
repeating units called nucleotides.
Each nucleotide is composed of a nitrogen-containing nucleobase—either , adenine (A), guanine (G), cytosine (C), or thymine (T)—as well as a monosaccharide sugar called deoxyribose and a phosphate group. - Nucleotides are summarized in the
table:

- Complementary nucleotides: adenine and thymine
are complements of each other, as are cytosine and guanine
bind to each other in DNA.

Complementary nucleotides: A & T, G & C -
Purine (C 5 H 4 N 4) and Pyrimidine (C 4 H 4
N 2) make up the two groups of nitrogenous bases, including
the two groups of nucleotide bases. Both purine
and pyrimidine are heterocyclic aromatic organic compound.
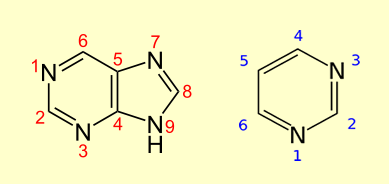
Purine (left) and Pyrimidine (right) - Replication: Replication begins in a genomic region called the replication origin (denoted oriC) and is performed by molecular copy machines called DNA polymerases. Locating oriC presents an important task not only for understanding how cells re,plicate but also for various biomedical problems.
- Computational Analysis: To find the replication origin, computational methods are much faster than experimental approaches; in addition, the results of many experiments cannot be interpreted without computational analysis.
- k-mer: In computational genomics, k-mers
refer to all the possible substrings of length k from a read
obtained through DNA Sequencing. The amount of k-mers possible
given a string of length, L, is L-k+1 whilst the
number of possible k-mers given n possibilities (4
in the case of DNA e.g. ACTG) is n^k.
K-mers are typically used during sequence assembly, but can also
be used in sequence alignment.

Subscribe to:
Comments (Atom)


