Here is my
on-going study notes at the entry point of this fascinating multi-dimensional
space - Bioinformatics.

- Genome: a full set of chromosomes; all the inheritable traits of an organism.
- DNA: Deoxyribonucleic acid (DNA) is a molecule that carries most of the genetic instructions used in the development, functioning and reproduction of all known living organisms and many viruses. Most DNA molecules consist of two biopolymer strands coiled around each other to form a double helix.
- Nucleic Acid: DNA (along with RNA) is a nucleic acid; alongside carbohydrates, lipids, proteins, and nucleic acids compose the four major macromolecules essential for all known forms of life.
- Nucleotide: DNA is a long polymer made from
repeating units called nucleotides.
Each nucleotide is composed of a nitrogen-containing nucleobase—either , adenine (A), guanine (G), cytosine (C), or thymine (T)—as well as a monosaccharide sugar called deoxyribose and a phosphate group. - Nucleotides are summarized in the
table:

- Complementary nucleotides: adenine and thymine
are complements of each other, as are cytosine and guanine
bind to each other in DNA.

Complementary nucleotides: A & T, G & C -
Purine (C 5 H 4 N 4) and Pyrimidine (C 4 H 4
N 2) make up the two groups of nitrogenous bases, including
the two groups of nucleotide bases. Both purine
and pyrimidine are heterocyclic aromatic organic compound.
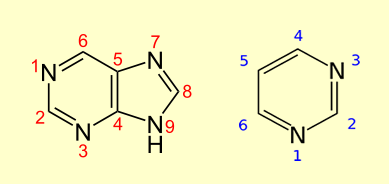
Purine (left) and Pyrimidine (right) - Replication: Replication begins in a genomic region called the replication origin (denoted oriC) and is performed by molecular copy machines called DNA polymerases. Locating oriC presents an important task not only for understanding how cells re,plicate but also for various biomedical problems.
- Computational Analysis: To find the replication origin, computational methods are much faster than experimental approaches; in addition, the results of many experiments cannot be interpreted without computational analysis.
- k-mer: In computational genomics, k-mers
refer to all the possible substrings of length k from a read
obtained through DNA Sequencing. The amount of k-mers possible
given a string of length, L, is L-k+1 whilst the
number of possible k-mers given n possibilities (4
in the case of DNA e.g. ACTG) is n^k.
K-mers are typically used during sequence assembly, but can also
be used in sequence alignment.

No comments:
Post a Comment